Skin Cancer Detection using CNN (VGG16) inculcated with CLAH and Gaussian Filter
Main Article Content
Abstract
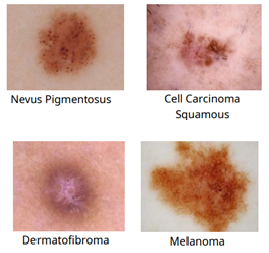
Many techniques related to image analysis have been proposed by researchers which are being used to detect a large number of diseases. These images are carefully analyzed by radiologists and doctors, and after careful interpretation, the results are obtained which finally help in making an appropriate diagnosis. This is a complicated and time consuming task, which requires high levels of concentration. Therefore, the experts who analyze the images mustn't suffer from fatigue or other common problems that can impair their performance. The present study attempts to reveal how a deep learning model using CNN with VGG16 is effective for the diagnosis and detection of skin cancer at its early stages. Therefore under the scheme, the 4000 images of raw skin cancer tissues are evaluated. The diagnostic model starts with pre-processing of images using the CLAHE along with the inculcation of the Gaussian filter. Thereafter, using hyper-parameter optimizer stochastic gradient descent, along with the effective learning rate 0.001, incorporating the training epochs of 50 nos. and pertaining the batch size 32 is formed. Consequently, as a result, the accuracy achieved is 99.70%, with a loss value of 0.0055%, a precision of 99.75%, a recall of 99.75%, and an f1-score of 99.50% respectively.
Article Details
References
Lee, S., Huang, H. and Zelen, M. (2004) “Early detection of disease and scheduling of screening examinations,” Statistical Methods in Medical Research, 13(6), pp. 443–456. Available at: https://doi.org/10.1191/0962280204sm377ra.
L. J. Loescher, PhD, RN, A. L. Howerter, PhD, K. M. Heslin, MPH, C. M. Azzolina, MPH, and M. L. Muramoto, MD, MPH, “A Survey of Licensed Massage Therapists’ Perceptions of Skin Cancer Prevention and Detection Activities,” International Journal of Therapeutic Massage & Bodywork: Research, Education, & Practice, vol. 11, no. 4, pp. 4–10, Dec. 2018, doi: https://doi.org/10.3822/ijtmb.v11i4.417.
R. Kaur, H. GholamHosseini, R. Sinha, and M. Lindén, “Melanoma Classification Using a Novel Deep Convolutional Neural Network with Dermoscopic Images,” Sensors, vol. 22, no. 3, p. 1134, Feb. 2022, doi: https://doi.org/10.3390/s22031134.
O. T. Jones, C. K. I. Ranmuthu, P. N. Hall, G. Funston, and F. M. Walter, “Recognising Skin Cancer in Primary Care,” Advances in Therapy, vol. 37, no. 1, pp. 603–616, Nov. 2019, doi: https://doi.org/10.1007/s12325-019-01130-1.
A. Esteva et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118, Jan. 2017, doi: https://doi.org/10.1038/nature21056.
Sunil Patil, Rakesh Saxena, Yogesh Pahariya. (2023). Performance Comparison of SRM, PMSM & BLDC Motor Drives via Experimentation in Laboratory for EV Application. International Journal of Intelligent Systems and Applications in Engineering, 11(2s), 405 –. Retrieved from https://ijisae.org/index.php/IJISAE/article/view/2736
G. P. Pfeifer, “Mechanisms of UV-induced mutations and skin cancer,” Genome Instability & Disease, Mar. 2020, doi: https://doi.org/10.1007/s42764-020-00009-8.
Mrs. Leena Rathi. (2014). Ancient Vedic Multiplication Based Optimized High Speed Arithmetic Logic . International Journal of New Practices in Management and Engineering, 3(03), 01 - 06. Retrieved from http://ijnpme.org/index.php/IJNPME/article/view/29
J. Ludzik, A. L. Becker, C. Lee, and A. Witkowski, “Augmenting pigmented lesion assay results with the three point dermoscopy checklist to improve pigmented lesion triage,” Skin Research and Technology, vol. 28, no. 6, pp. 877–879, Sep. 2022, doi: https://doi.org/10.1111/srt.13205.
N. B. Linsangan and J. J. Adtoon, “Skin Cancer Detection and Classification for Moles Using K-Nearest Neighbor Algorithm,” Proceedings of the 2018 5th International Conference on Bioinformatics Research and Applications - ICBRA ’18, 2018, doi: https://doi.org/10.1145/3309129.3309141.
Y. N. Fu’adah, N. C. Pratiwi, M. A. Pramudito, and N. Ibrahim, “Convolutional Neural Network (CNN) for Automatic Skin Cancer Classification System,” IOP Conference Series: Materials Science and Engineering, vol. 982, p. 012005, Dec. 2020, doi: https://doi.org/10.1088/1757-899x/982/1/012005. G. Eason, B. Noble, and I. N. Sneddon, “On certain integrals of Lipschitz-Hankel type involving products of Bessel functions,” Phil. Trans. Roy. Soc. London, vol. A247, pp. 529–551, April 1955. (references)
Simonyan, Karen and Andrew Zisserman. “Very Deep Convolutional Networks for Large-Scale Image Recognition.” CoRR abs/1409.1556 (2014): n. pag.
E. J. Topol, “High-performance medicine: the convergence of human and artificial intelligence,” Nature Medicine, vol. 25, no. 1, pp. 44–56, Jan. 2019, doi: https://doi.org/10.1038/s41591-018-0300-7.
Kadry, S., Rajinikanth, V., Raja, N.S.M. et al. "Evaluation of brain tumor using brain MRI with modified-moth-flame algorithm and Kapur’s thresholding: a study". Evol. Intel. 14, 1053–1063 (2021). https://doi.org/10.1007/s12065-020-00539-w
Howard, Andrew, et al. "MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications." ArXiv, 2017, https://doi.org/10.48550/arXiv.1704.04861. Accessed 15 Feb. 2023.
M. F. Alanazi et al., “Brain Tumor/Mass Classification Framework Using Magnetic-Resonance-Imaging-Based Isolated and Developed Transfer Deep-Learning Model,” Sensors, vol. 22, no. 1, p. 372, Jan. 2022, doi: https://doi.org/10.3390/s22010372.
M. M. Yamunadevi and S. S. Ranjani, “Retraction Note to: Efficient segmentation of the lung carcinoma by adaptive fuzzy–GLCM (AF-GLCM) with deep learning based classification,” Journal of Ambient Intelligence and Humanized Computing, Jul. 2022, doi: https://doi.org/10.1007/s12652-022-04267-0.
P. S. Ganesh, T. S. Kumar, M. Kumar, and Mr. S. R. Kumar, “Brain Tumor Detection and Classification Using Image Processing Techniques,” International Journal of Advanced Research in Science, Communication and Technology, pp. 32–38, Apr. 2021, doi: https://doi.org/10.48175/ijarsct-v4-i3-005.
Kshirsagar, D. P. R. ., Patil, D. N. N. ., & Makarand L., M. . (2022). User Profile Based on Spreading Activation Ontology Recommendation. Research Journal of Computer Systems and Engineering, 3(1), 73–77. Retrieved from https://technicaljournals.org/RJCSE/index.php/journal/article/view/45
A. W. Setiawan, "Effect of Color Enhancement on Early Detection of Skin Cancer using Convolutional Neural Network," 2020 IEEE International Conference on Informatics, IoT, and Enabling Technologies (ICIoT), Doha, Qatar, 2020, pp. 100-103, doi: 10.1109/ICIoT48696.2020.9089631.
Dr. K. Kanthamma, “Improved CLAHE Enhancement Technique for Underwater Images,” International Journal of Engineering Research and, vol. V9, no. 07, Jul. 2020, doi: https://doi.org/10.17577/ijertv9is070064.
Ming Zhang and Bahadir Gunturk, "A new image denoising method based on the bilateral filter," 2008 IEEE International Conference on Acoustics, Speech and Signal Processing, 2008, pp. 929-932, doi: 10.1109/ICASSP.2008.4517763.
Mohan Patro and M. Ranjan Patra, “A Novel Approach to Compute Confusion Matrix for Classification of n-Class Attributes with Feature Selection,” Transactions on Machine Learning and Artificial Intelligence, Apr. 2015, doi: https://doi.org/10.14738/tmlai.32.1108.
“Understanding Categorical Cross-Entropy Loss, Binary Cross-Entropy Loss, Softmax Loss, Logistic Loss, Focal Loss and all those confusing names,” Github.io, 2018. https://gombru.github.io/2018/05/23/cross_entropy_loss/
[1]S. Bera and V. K. Shrivastava, “Analysis of various optimizers on deep convolutional neural network model in the application of hyperspectral remote sensing image classification,” International Journal of Remote Sensing, vol. 41, no. 7, pp. 2664–2683, Dec. 2019, doi: https://doi.org/10.1080/01431161.2019.1694725.