A Review on Skin Disease Classification and Detection Using Deep Learning Techniques
Main Article Content
Abstract
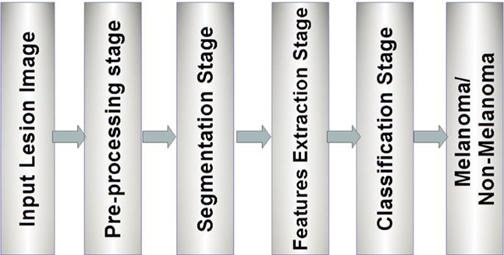
Skin cancer ranks among the most dangerous cancers. Skin cancers are commonly referred to as Melanoma. Melanoma is brought on by genetic faults or mutations on the skin, which are caused by Unrepaired Deoxyribonucleic Acid (DNA) in skin cells. It is essential to detect skin cancer in its infancy phase since it is more curable in its initial phases. Skin cancer typically progresses to other regions of the body. Owing to the disease's increased frequency, high mortality rate, and prohibitively high cost of medical treatments, early diagnosis of skin cancer signs is crucial. Due to the fact that how hazardous these disorders are, scholars have developed a number of early-detection techniques for melanoma. Lesion characteristics such as symmetry, colour, size, shape, and others are often utilised to detect skin cancer and distinguish benign skin cancer from melanoma. An in-depth investigation of deep learning techniques for melanoma's early detection is provided in this study. This study discusses the traditional feature extraction-based machine learning approaches for the segmentation and classification of skin lesions. Comparison-oriented research has been conducted to demonstrate the significance of various deep learning-based segmentation and classification approaches.
Article Details
References
Davis, K. E. (2020). Skin Cancer Rates Rising. Journal of the Dermatology Nurses' Association, 12(3), 113-114.
Le, D. N., Le, H. X., Ngo, L. T., & Ngo, H. T. (2020). Transfer learning with class-weighted and focal loss function for automatic skin cancer classification. arXiv preprint arXiv:2009.05977.
https://www.cancer.org/cancer/melanoma-skin- cancer/about/key-statistics.html
https://theprint.in/health/india-sees-high-number-of-skin- cancer-cases-in-north-northeast-regions-icmr-study- finds/741687/
https://www.uicc.org/news/global-cancer-data-globocan- 2018
Miller, K. D., Siegel, R. L., Lin, C. C., Mariotto, A. B., Kramer, J. L., Rowland, J. H., ... & Jemal, A. (2016). Cancer treatment and survivorship statistics, 2016. CA: a cancer journal for clinicians, 66(4), 271-289.
Goodall, G. J., & Wickramasinghe, V. O. (2021). RNA in cancer. Nature Reviews Cancer, 21(1), 22-36.
Nida, N., Shah, S. A., Ahmad, W., Faizi, M. I., & Anwar, S. random fields. Multimedia Tools and Applications, 1-21.
Pallua, N., & Kim, B. S. (2020). Reconstruction Principles of Localized Non-melanoma Skin Cancer and Cutaneous Melanoma. In Non-Melanoma Skin Cancer and Cutaneous Melanoma (pp. 243-263). Springer, Cham.
Feller, L., Khammissa, R. A. G., Kramer, B., Altini, M., & Lemmer, J. (2016). Basal cell carcinoma, squamous cell carcinoma and melanoma of the head and face. Head & face medicine, 12(1), 1-7.
Howell, J. Y., & Ramsey, M. L. (2022). Squamous cell skin cancer. In StatPearls [Internet]. StatPearls Publishing.
Chase, T., Cham, K. E., & Cham, B. E. (2020). Curaderm, the Long-Awaited Breakthrough for Basal Cell Carcinoma. International Journal of Clinical Medicine, 11(10), 579.
Johnson, D. E., Burtness, B., Leemans, C. R., Lui, V. W. Y., Bauman, J. E., & Grandis, J. R. (2020). Head and neck squamous cell carcinoma. Nature reviews Disease primers, 6(1), 1-22.
Zaidi, M. R., Fisher, D. E., & Rizos, H. (2020). Biology of melanocytes and primary melanoma. Cutaneous Melanoma, 3-40.
Kutzner, H., Jutzi, T. B., Krahl, D., Krieghoff?Henning, E. I., Heppt, M. V., Hekler, A., ... & Brinker, T. J. (2020). Overdiagnosis of melanoma–causes, consequences and solutions. JDDG: Journal der Deutschen Dermatologischen Gesellschaft, 18(11), 1236-1243.
Majumder, S., & Ullah, M. A. (2019). Feature extraction from dermoscopy images for melanoma diagnosis. SN Applied Sciences, 1(7), 1-11.
Roberts, D. L. L., Anstey, A. V., Barlow, R. J., Cox, N. H., British Association of Dermatologists, Bishop, J. N., ... & Kirkham, N. (2002). UK guidelines for the management of cutaneous melanoma. British Journal of dermatology, 146(1), 7-17.
Treatment, M. (2018). Health Professional Version.(nd) PDQ Adult Treatment Editorial Board. PDQ Cancer Information Summaries [Internet]. Bethesda (MD): National Cancer Institute (US).
Balch, C. M., Buzaid, A. C., Atkins, M. B., Cascinelli, N., Coit, D. G., Fleming, I. D., ... & AJCC Melanoma Staging Committee. (2000). A new American Joint Committee on Cancer staging system for cutaneous melanoma. Cancer, 88(6), 1484-1491.
Sabir, F., Barani, M., Rahdar, A., Bilal, M., & Nadeem, M. (2021). How to face skin cancer with nanomaterials: A review. Biointerface Res. Appl. Chem, 11, 11931-11955.
Pashazadeh, A., Boese, A., & Friebe, M. (2019). Radiation therapy techniques in the treatment of skin cancer: an overview of the current status and outlook. Journal of Dermatological Treatment.
Rosenberg, A. R., Tabacchi, M., Ngo, K. H., Wallendorf, M., Rosman, I. S., Cornelius, L. A., & Demehri, S. (2019). Skin cancer precursor immunotherapy for squamous cell carcinoma prevention. JCI insight, 4(6).
Siegel RL, Miller KD and Jemal A (2018) Cancer statistics. CA: A Cancer Journal for Clinicians 68: 7–30
Afza, F., Khan, M. A., Sharif, M., & Rehman, A. (2019). Microscopic skin laceration segmentation and classification: A framework of statistical normal distribution and optimal feature selection. Microscopy research and technique, 82(9), 1471-1488.
Kuzmina, I., Diebele, I., Jakovels, D., Spigulis, J., Valeine, L., Kapostinsh, J., & Berzina, A. (2011). Towards noncontact skin melanoma selection by multispectral imaging analysis. Journal of Biomedical optics, 16(6), 060502.
Querleux, B., Guillot, G., Ginéfri, J. C., Poirier?Quinot, M., & Darrasse, L. (2019). Quantitative Magnetic Resonance Imaging of the Skin: In Vitro and In Vivo Applications. Imaging Technologies and Transdermal Delivery in Skin Disorders, 341-369.
Aumann, S., Donner, S., Fischer, J., & Müller, F. (2019). Optical coherence tomography (OCT): principle and technical realization. High Resolution Imaging in Microscopy and Ophthalmology, 59-85.
Catalano, O., Roldán, F. A., Varelli, C., Bard, R., Corvino, A., & Wortsman, X. (2019). Skin cancer: findings and role of high-resolution ultrasound. Journal of Ultrasound, 22(4), 423-431
Ilie, M. A., Caruntu, C., Lupu, M., Lixandru, D., Tampa, M., Georgescu, S. R., ... & Boda, D. (2019). Current and future applications of confocal laser scanning microscopy imaging in skin oncology. Oncology letters, 17(5), 4102-4111.
Weber, P., Tschandl, P., Sinz, C., & Kittler, H. (2018). Dermatoscopy of neoplastic skin lesions: recent advances, updates, and revisions. Current Treatment Options in Oncology, 19(11), 1-17.
Pacheco, A. G., & Krohling, R. A. (2020). The impact of patient clinical information on automated skin cancer detection. Computers in biology and medicine, 116, 103545.
Vocaturo, E., Zumpano, E., & Veltri, P. (2018, December). Image pre-processing in computer vision systems for melanoma detection. In 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (pp. 2117-2124). IEEE.
Siegel, R. L., Miller, K. D., Fuchs, H. E., & Jemal, A. (2021). Cancer statistics, 2021. Ca Cancer J Clin, 71(1), 7-33.
Razzak, I., Shoukat, G., Naz, S., & Khan, T. M. (2020, July). Skin lesion analysis toward accurate detection of melanoma using multistage fully connected residual network. In 2020 International Joint Conference on Neural Networks (IJCNN) (pp. 1-8). IEEE.
Anand, V., Gupta, S., Nayak, S. R., Koundal, D., Prakash, D., & Verma, K. D. (2022). An automated deep learning models for classification of skin disease using Dermoscopy images: a comprehensive study. Multimedia Tools and Applications, 1-23.
Dildar, M., Akram, S., Irfan, M., Khan, H. U., Ramzan, M., Mahmood, A. R., ... & Mahnashi, M. H. (2021). Skin cancer detection: a review using deep learning techniques. International journal of environmental research and public health, 18(10), 5479.
Nahata, H., & Singh, S. P. (2020). Deep learning solutions for skin cancer detection and diagnosis. In Machine Learning with Health Care Perspective (pp. 159-182). Springer, Cham.
Nassif, A. B., Shahin, I., Attili, I., Azzeh, M., & Shaalan, K. (2019). Speech recognition using deep neural networks: A systematic review. IEEE access, 7, 19143-19165.
Liu, W., Wang, Z., Liu, X., Zeng, N., Liu, Y., & Alsaadi, F. (2017). A survey of deep neural network architectures and their applications. Neurocomputing, 234, 11-26.
Siddesha, K., Jayaramaiah, G. V., & Singh, C. (2022). A novel deep reinforcement learning scheme for task scheduling in cloud computing. Cluster Computing, 1-18.
Min, S., Lee, B., & Yoon, S. (2017). Deep learning in bioinformatics. Briefings in bioinformatics, 18(5), 851-869.
Tajjour, S., Garg, S., Chandel, S. S., & Sharma, D. (2022). A novel hybrid artificial neural network technique for the early skin cancer diagnosis using color space conversions of original images. International Journal of Imaging Systems and Technology.
Al Mamun, M., & Uddin, M. S. (2021). A Comparative Study among Segmentation Techniques for Skin Disease Detection Systems. In Proceedings of International Conference on Trends in Computational and Cognitive
Mishra, N. K., & Celebi, M. E. (2016). An overview of melanoma detection in dermoscopy images using image processing and machine learning. arXiv preprint arXiv:1601.07843.
Li, W.; Raj, A.N.J.; Tjahjadi, T.; Zhuang, Z. Digital hair removal by deep learning for skin lesion segmentation. Pattern Recognit. 2021, 117, 107994.
Guarracino, M.R.; Maddalena, L. SDI+: A novel algorithm for segmenting dermoscopic images. IEEE J. Biomed. Health Inform. 2019, 23, 481–488.
Jahanifar, M.; Tajeddin, N.Z.; Gooya, A.; Asl, B.M. Segmentation of lesions in dermoscopy images using saliency map and contour propagation. arXiv 2017, arXiv:1703.00087v2
Nida, N.; Irtaza, A.; Javed, A.; Yousaf, M.H.; Mahmood, M.T. Melanoma lesion detection and segmentation using deep region based convolutional neural network and fuzzy C-means clustering. Int. J. Med. Inform. 2019, 124, 37–48.
Pennisi, A.; Bloisi, D.D.; Nardi, D.; Giampetruzzi, A.R.; Mondino, C.; Facchiano, A. Skin lesion image segmentation using Delaunay Triangulation for melanoma detection. Comput. Med. Imaging Graph. 2016, 52, 89–103.
Guarracino, M.R.; Maddalena, L. SDI+: A novel algorithm for segmenting dermoscopic images. IEEE J. Biomed. Health Inform. 2019, 23, 481–488.
Ünver, H.M.; Ayan, E. Skin lesion segmentation in dermoscopic images with combination of YOLO and grabcut algorithm. Diagnostics 2019, 9, 72
Hu, K.; Liu, S.; Zhang, Y.; Cao, C.; Xiao, F.; Huang, W.; Gao, X. Automatic segmentation of dermoscopy images using saliency combined with adaptive thresholding based on wavelet transform. Multimed. Tools Appl. 2020, 79, 14625–14642
Garnavi, R.; Aldeen, M.; Celebi, M.E.; Varigos, G.; Finch, S. Border detection in dermoscopy images using hybrid thresholding on optimized color channels. Comput. Med. Imaging Graph. 2011, 35, 105–115.
Jahanifar, M.; Tajeddin, N.Z.; Gooya, A.; Asl, B.M. Segmentation of lesions in dermoscopy images using saliency map and contour propagation. arXiv 2017, arXiv:1703.00087v2.
Joseph, S., & Olugbara, O. O. (2022). Preprocessing effects on performance of skin lesion saliency segmentation. Diagnostics, 12(2), 344.
Samsudin, S. S., Arof, H., Harun, S. W., Abdul Wahab, A. W., & Idris, M. Y. I. (2022). Skin lesion classification using multi-resolution empirical mode decomposition and local binary pattern. Plos one, 17(9), e0274896.
Sarker, M. M. K., Rashwan, H. A., Akram, F., Singh, V. K., Banu, S. F., Chowdhury, F. U., ... & Abdel-Nasser, M. (2021). SLSNet: Skin lesion segmentation using a lightweight generative adversarial network. Expert Systems with Applications, 183, 115433.
Kimori, Y. (2022). A Morphological Image Preprocessing Method Based on the Geometrical Shape of Lesions to Improve the Lesion Recognition Performance of Convolutional Neural Networks. IEEE Access, 10, 70919- 70936.
Hossen, E. (2021). Design a Convolutional Neural Network for Melanocytic Skin Lesion Identification (Doctoral dissertation, Texas A&M University-Commerce).
Abduljaleel, H. K. (2019). Deep CNN based skin lesion image denoising and segmentation using active contour method. Engineering and Technology Journal, 37(11 Part A).
Guo, Y., Ashour, A. S., & Smarandache, F. (2018). A novel skin lesion detection approach using neutrosophic clustering and adaptive region growing in dermoscopy images. Symmetry, 10(4), 119.
Devi, S. S., Laskar, R. H., & Singh, N. H. (2020). Fuzzy C- means clustering with histogram based cluster selection for skin lesion segmentation using non-dermoscopic images.
J. Long, E. Shelhamer, and T. Darrell, ‘‘Fully convolutional networks for semantic segmentation,’’ in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2015, pp. 3431–3440.
O. Ronneberger, P. Fischer, and T. Brox, ‘‘U-Net: Convolutional networks for biomedical image segmentation,’’ in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Cham, Switzerland: Springer, 2015, pp. 234–241
V. Badrinarayanan, A. Kendall, and R. Cipolla, ‘‘SegNet: A deep convolutional encoder-decoder architecture for image segmentation,’’ IEEE Trans. Pattern Anal. Mach. Intell., vol. 39, no. 12, pp. 2481–2495, Dec. 2017
L. C. Chen, G. Papandreou, and I. Kokkinos, ‘‘DeepLab: Semantic image segmentation with deep convolutional nets, Atrous convolution, and fully connected CRFs,’’ IEEE Trans. Pattern Anal. Mach. Intell., vol. 40, no. 4, pp. 834– 848, Jun. 2016
Y. Xie, J. Zhang, Y. Xia, and C. Shen, ‘‘A mutual bootstrapping model for automated skin lesion segmentation and classification,’’ IEEE Trans. Med. Imag., vol. 39, no. 7, pp. 2482–2493, Dec. 2020.
K.-K. Maninis, S. Caelles, J. Pont-Tuset, and L. Van Gool, ‘‘Deep extreme cut: From extreme points to object segmentation,’’ in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., Jun. 2018, pp. 616–625.
A. Kumar, G. Hamarneh, and M. S. Drew, ‘‘Illumination-based transformations improve skin lesion segmentation in dermoscopic images,’’ in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. Workshops (CVPRW), Jun. 2020, pp. 728–729.
H. Wu, S. Chen, G. Chen, W. Wang, B. Lei, and Z. Wen, ‘‘FAT-Net: Feature adaptive transformers for automated skin lesion segmentation,’’ Med. Image Anal., vol. 76, Feb. 2022, Art. no. 102327
W.-S. Lai, J.-B. Huang, N. Ahuja, and M.-H. Yang, ‘‘Deep Laplacian pyramid networks for fast and accurate super- resolution,’’ in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jul. 2017, pp. 624–632.
R. Azad, M. Asadi-Aghbolaghi, M. Fathy, and S. Escalera, ‘‘Attention deeplabv3+: Multi-level context attention mechanism for skin lesion segmentation,’’ in Proc. Eur. Conf. Comput. Vis. Cham, Switzerland: Springer, 2020, pp. 251–266.
R. Azad, M. Asadi-Aghbolaghi, M. Fathy, and S. Escalera, ‘‘Bi-directional ConvLSTM U-Net with Densley connected convolutions,’’ in Proc. IEEE/CVF Int. Conf. Comput. Vis. Workshop (ICCVW), Oct. 2019, pp. 1–10
L. Liu, Y. Y. Tsui, and M. Mandal, ‘‘Skin lesion segmentation using deep learning with auxiliary task,’’ J. Imag., vol. 7, no. 4, p. 67, Apr. 2021.
D. Dai, C. Dong, S. Xu, Q. Yan, Z. Li, C. Zhang, and N. Luo, ‘‘Ms RED: A novel multi-scale residual encoding and decoding network for skin lesion segmentation,’’ Med. Image Anal., vol. 75, Jan. 2022, Art. no. 102293
Bi, L.; Kim, J.; Ahn, E.; Kumar, A.; Feng, D.; Fulham, M. Step-wise integration of deep class-specific learning for dermoscopic image segmentation. Pattern Recognit. 2019, 85, 78–89
Drozdzal, M.; Vorontsov, E.; Chartrand, G.; Kadoury, S.; Pal, C. The Importance of Skip Connections in Biomedical Image Segmentation. arXiv 2016, arXiv:1608.04117.
Wei, Z.; Song, H.; Chen, L.; Li, Q.; Han, G. Attention- Based DenseUnet Network With Adversarial Training for Skin Lesion Segmentation. IEEE Access 2019, 7, 136616– 136629.
Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net Architecture for Multimodal Biomedical Image Segmentation. Neural Netw. 2020, 121, 74–87
Kaul, C.; Manandhar, S.; Pears, N. Focusnet: An Attention- Based Fully Convolutional Network for Medical Image Segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 455–458
Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.-S. CBAM: Convolutional Block Attention Module. arXiv 2018, arXiv:1807.06521v2
Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and- Excitation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 2011–2023
Jaderberg, M.; Simonyan, K.; Zisserman, A.; Kavukcuoglu, K. Spatial Transformer Networks. arXiv 2015, arXiv:1506.02025.
Alencar, F.E.S.; Lopes, D.C.; Neto, F.M.M. Development of a System Classification of Images Dermoscopic for Mobile Devices. IEEE Lat. Am. Trans. 2016, 14, 325–330
Kasmi, R.; Mokrani, K. Classification of malignant melanoma and benign skin lesions: Implementation of automatic ABCD rule. IET Image Process. 2016, 10, 448–455. [CrossRef]
Sáez, A.; Sánchez-Monedero, J.; Gutiérrez, P.A.; Hervás- Martínez, C. Machine Learning Methods for Binary and Multiclass Classification of Melanoma Thickness From Dermoscopic Images. IEEE Trans. Med. Imaging 2016, 35, 1036–1045.
Ma, Z.; Tavares, J.M.R.S. A Novel Approach to Segment Skin Lesions in Dermoscopic Images Based on a Deformable Model. IEEE J. Biomed. Health Inform. 2016, 20, 615–623.
Oliveira, R.B.; Marranghello, N.; Pereira, A.S.; Tavares, J.M.R.S. A computational approach for detecting pigmented skin lesions in macroscopic images. Expert Syst. Appl. 2016, 61, 53–63.
Inácio, D.F.; Célio, V.N.; Vilanova, G.D.; Conceição, M.M.; Fábio, G.; Minoro, A.J.; Tavares, P.M.; Landulfo, S. Paraconsistent analysis network applied in the treatment of Raman spectroscopy data to support medical diagnosis of skin cancer. Med. Biol. Eng. Comput. 2016, 54, 1453–1467.
Pennisi, A.; Bloisi, D.D.; Nardi, D.A.; Giampetruzzi, R.; Mondino, C.; Facchiano, A. Skin lesion image segmentation using Delaunay Triangulation for melanoma detection. Comput. Med. Imaging Graph. 2016, 52, 89–103.
Prof. Naveen Jain. (2013). FPGA Implementation of Hardware Architecture for H264/AV Codec Standards. International Journal of New Practices in Management and Engineering, 2(01), 01 - 07. Retrieved from http://ijnpme.org/index.php/IJNPME/article/view/11
Noroozi, N.; Zakerolhosseini, A. Differential diagnosis of squamous cell carcinoma in situ using skin histopathological images. Comput. Biol. Med. 2016, 70, 23–39.
Xie, F.; Fan, H.; Li, Y.; Jiang, Z.; Meng, R.; Bovik, A. Melanoma Classification on Dermoscopy Images Using a Neural Network Ensemble Model. IEEE Trans. Med. Imaging 2017, 36, 849–858.
Dhabliya, D. (2021). An Integrated Optimization Model for Plant Diseases Prediction with Machine Learning Model . Machine Learning Applications in Engineering Education and Management, 1(2), 21–26. Retrieved from http://yashikajournals.com/index.php/mlaeem/article/view/15
Sadri, A.R.; Azarianpour, S.; Zekri, M.; Celebi, M.E.; Sadri, S. WN-based approach to melanoma diagnosis from dermoscopy images. IET Image Process. 2017, 11, 475–482.
Hamed, K.; Khaki, S.A.; Mohammad, A.; Jahed, M.; Ali, H. Nonlinear Analysis of the Contour Boundary Irregularity of Skin Lesion Using Lyapunov Exponent and K-S Entropy. J. Med. Biol. Eng. 2017, 37, 409–419.
Dalila, F.; Zohra, A.; Reda, K.; Hocine, C. Segmentation and classification of melanoma and benign skin lesions. Optik 2017, 140, 749–761.
Ma, Z.; Tavares, J.M.R.S. Effective features to classify skin lesions in dermoscopic images. Expert Syst. Appl. 2017, 84, 92–101.
Alfed, N.; Khelifi, F. Bagged textural and color features for melanoma skin cancer detection in dermoscopic and standard images. Expert Syst. Appl. 2017, 90, 101–110.
Oliveira, R.B.; Pereira, A.S.; Tavares, J.M.R.S. Skin lesion computational diagnosis of dermoscopic images: Ensemble models based on input feature manipulation. Comput. Methods Programs Biomed. 2017, 149, 43–53.
Przystalski, K.; Ogorza?ek, M.J. Multispectral skin patterns analysis using fractal methods. Expert Syst. Appl. 2017, 88, 318–326.
Jaisakthi, S.M.; Mirunalini, P.; Aravindan, C. Automated skin lesion segmentation of dermoscopic images using GrabCut and k-means algorithms. IET Comput. Vis. 2018, 12, 1088–1095.
Do, T.; Hoang, T.; Pomponiu, V.; Zhou, Y.; Chen, Z.; Cheung, N.; Koh, D.; Tan, A.; Tan, S. Accessible Melanoma Detection Using Smartphones and Mobile Image Analysis. IEEE Trans. Multimed. 2018, 20, 2849–2864. [CrossRef]
Adjed, F.; Gardezi, S.J.S.; Ababsa, F.; Faye, I.; Dass, S.C. Fusion of structural and textural features for melanoma recognition. IET Comput. Vis. 2018, 12, 185–195. [CrossRef]
Faris, W. F. . (2020). Cataract Eye Detection Using Deep Learning Based Feature Extraction with Classification. Research Journal of Computer Systems and Engineering, 1(2), 20:25. Retrieved from https://technicaljournals.org/RJCSE/index.php/journal/article/view/7
Hosseinzadeh, H. Automated skin lesion division utilizing Gabor filters based on shark smell optimizing method. Evol. Syst. 2018, 11, 1–10.
Tan, T.Y.; Zhang, L.; Neoh, S.C.; Lim, C.P. Intelligent skin cancer detection using enhanced particle swarm optimization. Knowl. Based Syst. 2018, 158, 118–135.
Tajeddin, N.Z.; Asl, B.M. Melanoma recognition in dermoscopy images using lesion’s peripheral region information. Comput. Methods Programs Biomed. 2018, 163, 143–153.
Filho, P.P.R.; Peixoto, S.A.; Nóbrega, R.V.M.; Hemanth, D.J.; Medeiros, A.G.; Sangaiah, A.K.; Albuquerque, V.H.C. Automatic histologically-closer classification of skin lesions. Comput. Med. Imaging Graph. 2018, 68, 40–54.
Peñaranda, F.; Naranjo, V.; Lloyd, G.; Kastl, L.; Kemper, B.; Schnekenburger, J.; Nallala, J.; Stone, N. Discrimination of skin cancer cells using Fourier transform infrared spectroscopy. Comput. Biol. Med. 2018, 100, 50–61.
Wahba, M.A.; Ashour, A.S.; Guo, Y.; Napoleon, S.A.; Elnaby, M.M.A. A novel cumulative level difference mean based GLDM and modified ABCD features ranked using eigenvector centrality approach for four skin lesion types classification. Comput. Methods Programs Biomed. 2018, 165, 163–174.
Zakeri, A.; Hokmabadi, A. Improvement in the diagnosis of melanoma and dysplastic lesions by introducing ABCD- PDT features and a hybrid classifier. Biocybern. Biomed. Eng. 2018, 38, 456–466
Sánchez-Monedero, J.; Pérez-Ortiz, M.; Sáez, A.; Gutiérrez, P.A.; Hervás-Martínez, C. Partial order label decomposition approaches for melanoma diagnosis. Appl. Soft Comput. 2018, 64, 341–355.
Hagerty, J.R.; Stanley, R.J.; Almubarak, H.A.; Lama, N.; Kasmi, R.; Guo, P.; Drugge, R.J.; Rabinovitz, H.S.; Oliviero, M.; Stoecker, W.V. Deep Learning and Handcrafted Method Fusion: Higher Diagnostic Accuracy for Melanoma Dermoscopy Images. IEEE J. Biomed. Health Inform. 2019, 23, 1385–1391.
Sarkar, R.; Chatterjee, C.C.; Hazra, A. Diagnosis of melanoma from dermoscopic images using a deep depthwise separable residual convolutional network. IET Image Process. 2019, 13, 2130–2142.
Zhang, J.; Xie, Y.; Xia, Y.; Shen, C. Attention Residual Learning for Skin Lesion Classification. IEEE Trans. Med. Imaging 2019, 38, 2092–2103.
Albahar, M.A. Skin Lesion Classification Using Convolutional Neural Network With Novel Regularizer. IEEE Access 2019, 7, 38306–38313.
González-Díaz, I. DermaKNet: Incorporating the Knowledge of Dermatologists to Convolutional Neural Networks for Skin Lesion Diagnosis. IEEE J. Biomed. Health Inform. 2019, 23, 547–559.
Flores, A., Silva, A., López, L., Rodriguez, A., & María, K. Machine Learning-Enabled Early Warning Systems for Engineering Student Retention. Kuwait Journal of Machine Learning, 1(1). Retrieved from http://kuwaitjournals.com/index.php/kjml/article/view/106
Kawahara, J.; Daneshvar, S.; Argenziano, G.; Hamarneh, G. Seven-Point Checklist and Skin Lesion Classification Using Multitask Multimodal Neural Nets. IEEE J. Biomed. Health Inform. 2019, 23, 538–546.
Yu, Z.;Jiang, X.; Zhou, F.; Qin, J.; Ni, D.; Chen, S.; Lei, B.; Wang, T. Melanoma Recognition in Dermoscopy Images via Aggregated Deep Convolutional Features. IEEE Trans. Biomed. Eng. 2019, 66, 1006–1016.
Dorj, U.; Lee, K.; Choi, J.; Lee, M. The skin cancer classification using deep convolutional neural network. Multimed. Tools Appl. 2018, 77, 9909–9924.
Gavrilov, D.A.; Melerzanov, A.V.; Shchelkunov, N.N.; Zakirov, E.I. Use of Neural Network-Based Deep Learning Techniques for the Diagnostics of Skin Diseases. Biomed. Eng. 2019, 52, 348–352.
Chang Lee, Deep Learning for Speech Recognition in Intelligent Assistants , Machine Learning Applications Conference Proceedings, Vol 1 2021.
Chen, M.; Zhou, P.; Wu, D.; Hu, L.; Hassan, M.M.; Alamri, A. AI-Skin: Skin disease recognition based on self- learning and wide data collection through a closed-loop framework. Inf. Fusion 2020, 54, 1–9.
Tan, T.Y.; Zhang, L.; Lim, C.P. Adaptive melanoma diagnosis using evolving clustering, ensemble and deep neural networks. Knowl. Based Syst. 2020, 187, 104807.
Khan, M.A.; Sharif, M.; Akram, T.; Damaševi?cius, R.; Maskeliunas, R. Skin lesion segmentation and multiclass classification using ¯ deep learning features and improved moth flame optimization. Diagnostics 2021, 11, 811
Tschandl, P.; Sinz, C.; Kittler, H. Domain-specific classification-pretrained fully convolutional network encoders for skin lesion segmentation. Comput. Biol. Med. 2019, 104, 111–116.
Vasconcelos, F.F.X.; Medeiros, A.G.; Peixoto, S.A.; Filho, P.P.R. Automatic skin lesions segmentation based on a new morphological approach via geodesic active contour. Cogn. Syst. Res. 2019, 55, 44–59.
Kwasigroch, A.; Grochowski, M.; Miko?ajczyk, A. Neural Architecture Search for Skin Lesion Classification. IEEE Access 2020, 8, 9061–9071.
Adegun, A.A.; Viriri, S. Deep Learning-Based System for Automatic Melanoma Detection. IEEE Access 2020, 8, 7160–7172.
Song, L.; Lin, J.P.; Wang, Z.J.; Wang, H. An End-to-end Multi-task Deep Learning Framework for Skin Lesion Analysis. IEEE J. Biomed. Health Inform. 2020.
Wei, L.; Ding, K.; Hu, H. Automatic Skin Cancer Detection in Dermoscopy Images Based on Ensemble Lightweight Deep Learning Network. IEEE Access 2020, 8, 99633–99647.
Mahbod, A.; Schaefer, G.; Wang, C.; Dorffner, G.; Ecker, R.; Ellinger, I. Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput. Methods Programs Biomed. 2020, 193, 105475.
Nagesh Appe, S. R. ., Arulselvi, G. ., & Balaji, G. . (2023). Tomato Ripeness Detection and Classification using VGG based CNN Models. International Journal of Intelligent Systems and Applications in Engineering, 11(1), 296–302. Retrieved from https://ijisae.org/index.php/IJISAE/article/view/2538
Hameed, N.; Shabut, A.M.; Ghosh, M.K.; Hossain, M.A. Multi-class multi-level classification algorithm for skin lesions classification using machine learning techniques. Expert Syst. Appl. 2020, 141, 112961.
Zhang, N.; Cai, Y.; Wang, Y.; Tian, Y.; Wang, X.; Badami, B. Skin cancer diagnosis based on optimized convolutional neural network. Artif. Intell. Med. 2020, 102, 101756.
Hasan, K.; Dahal, L.; Samarakoon, P.N.; Tushar, F.I.; Martí, R. DSNet: Automatic dermoscopic skin lesion segmentation. Comput. Biol. Med. 2020, 120, 103738.